Knowledge-Driven Online Multimodal Automated Phenotyping System (KOMAP)
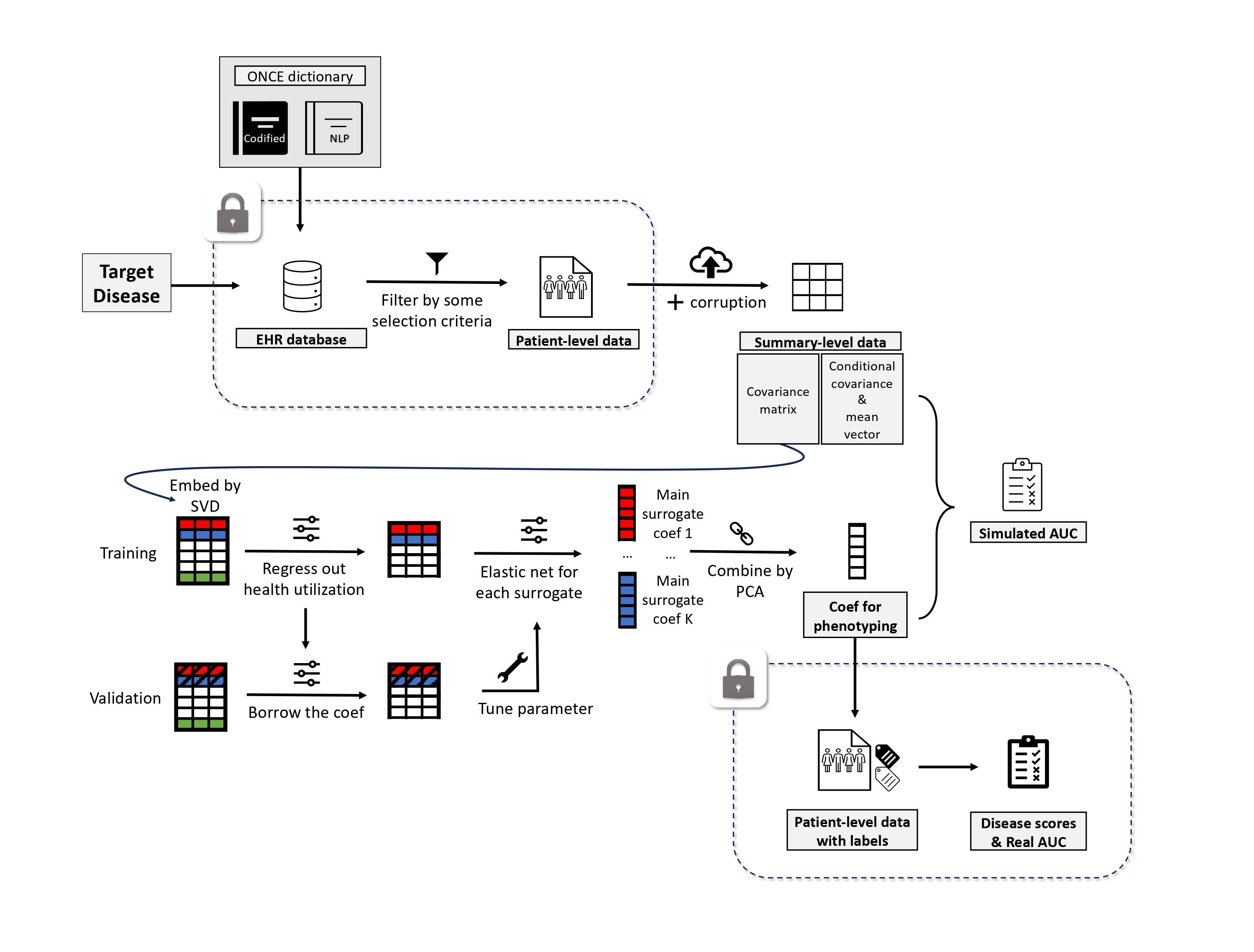
About KOMAP
The KOMAP pipeline comprises two key components:
Feature Selection: using the Online Narrative and Codified feature Search engine (ONCE), powered by multi-source knowledge graph representation and illustrated in the ONCE webapp
Online Phenotyping Algorithm Training and Validation: KOMAP can train a multimodal phenotyping algorithm fully online based on a user-supplied summary of the feature matrix. KOMAP also contains an online evaluation system to approximate evaluation metrics based on additional summary statistics derived from a validation set of labeled data.
How does it work?
With a given set of selected features, including a set of main surrogate features which can be indicated by ONCE and a healthcare utilization measure, the training of KOMAP contains three steps:
- Normalizing the main surrogates with the utilization
- Denoising via regression on each main surrogate
- Combining the derived risk scores of different surrogates
The only requirement for training is the empirical covariance matrix, free of any patient-level data.
The key working assumption behind the proposed evaluation algorithm is that all the features given the label approximately follow a Gaussian distribution. With this assumption, the ROC curve of the predicted score is uniquely determined by the conditional mean vectors and conditional covariance matrices.
To read more about KOMAP and our paired feature selection app, ONCE, you can view our paper on medRxiv.
You can also view our R package on github for additional information on formatting and creating the required inputs for the web app.
Quick Start Guide
Step 0 (Optional): Identify a list of features related to your disease of interest using ONCE
Step 1 - Create Input: Upload the training and validation covariance matrices with corrupted main surrogates and upload your dictionary connecting variable names to their descriptions
Step 2 - Name Inputs: Specify column names for main surrogate feature(s) and the healthcare utility corresponding to the disease
Step 3 (Optional) - Add Labeled Input: With label data, upload prevalence, conditional mean vectors and conditional covariance matrices;
Step 4 - Train and Validate: Click the “GO KOMAP” button and you are ready to go!
Model inputs:
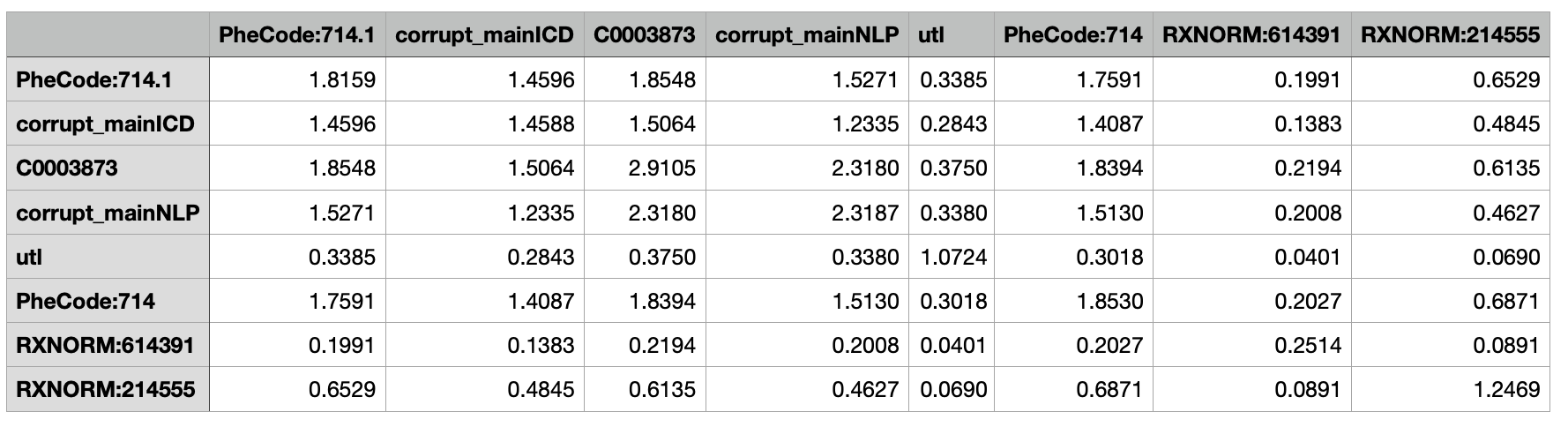
Training covariance matrix
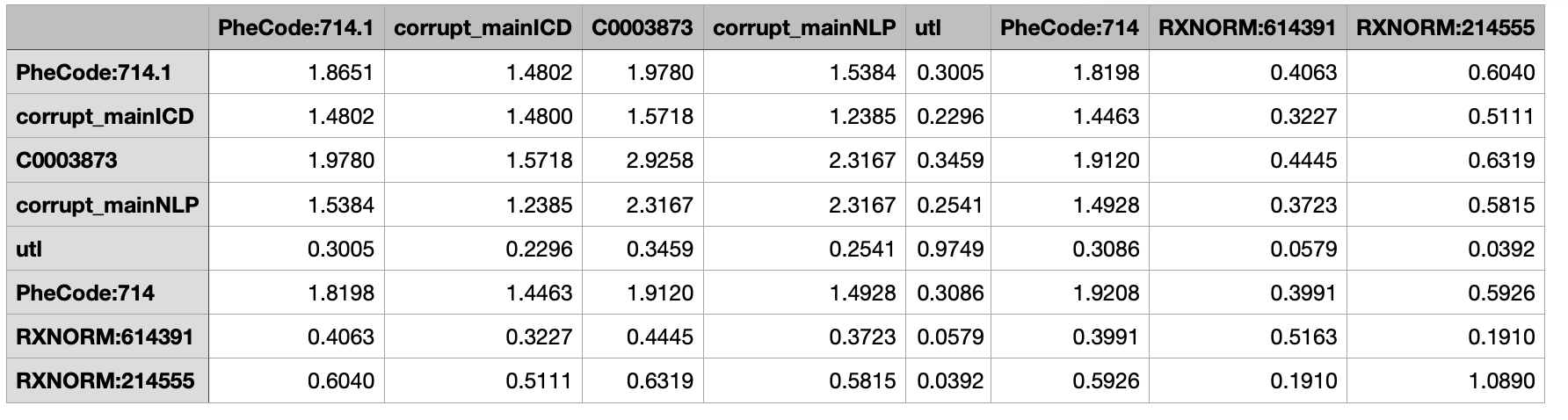
Validation covariance matrix
Notice!
- Training and validation covariance matrices must have the same set of concpets as their column names and row names.
- There must exist at least one main surrogate and its corrupted version in each covariance matrix.
- Corrupted surrogate is generated by replacing 20% of surrogate by its mean.
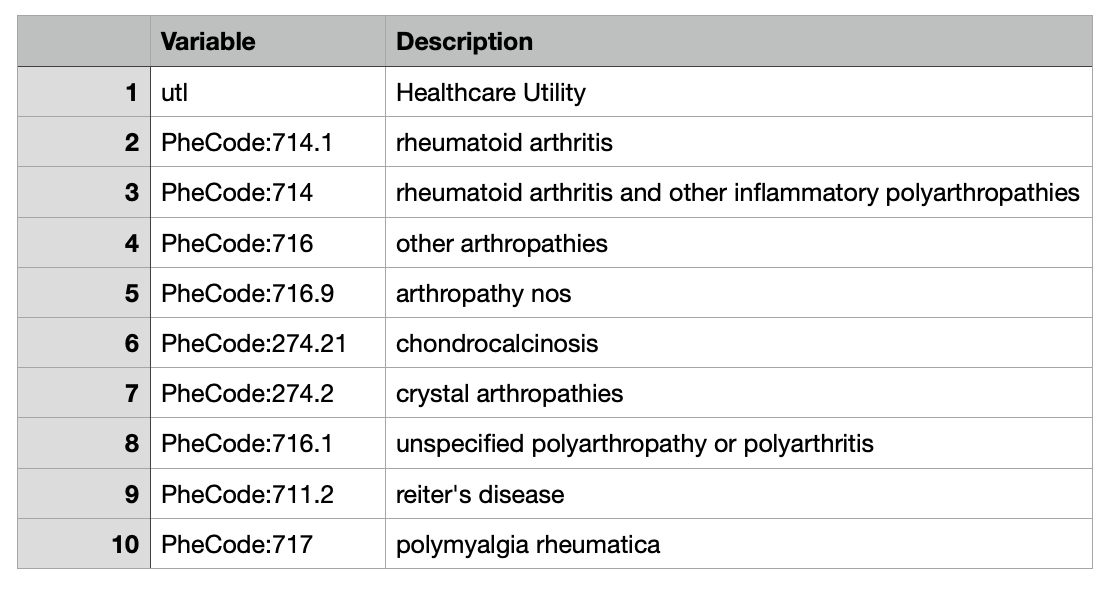
Dictionary
Features
- Specify the number of surrogates you want to fit.
- Identify the name of each surrogate as well as its corrupted version.
- Identify the name of the healthcare utilization score.
Conditional summary data
Wrap up the following summary-level data into one excel file:
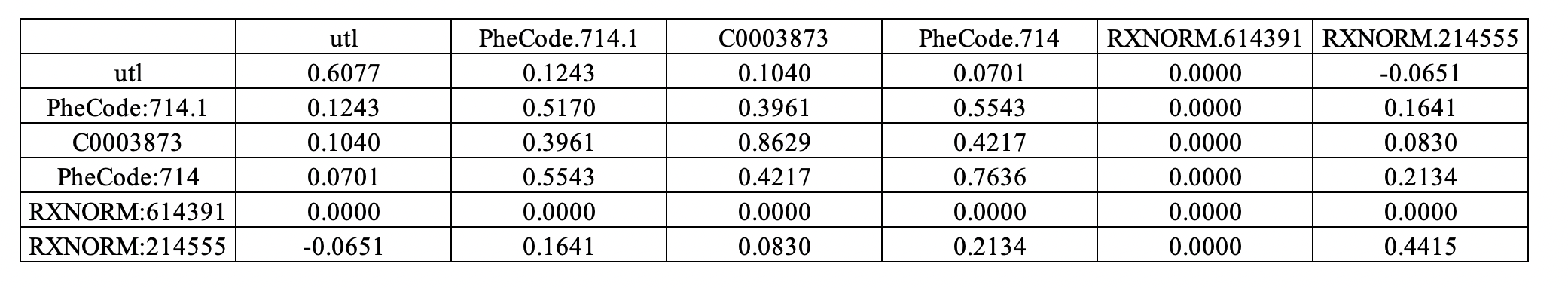
- Sheet 1: Conditional covariance matrix among patients with negative disease status.
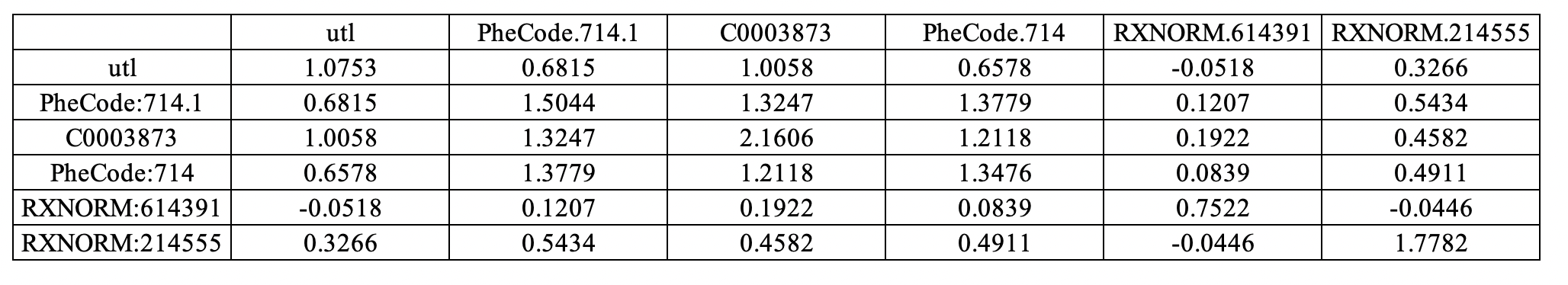
- Sheet 2: Conditional covariance matrix among patients with positive disease status.
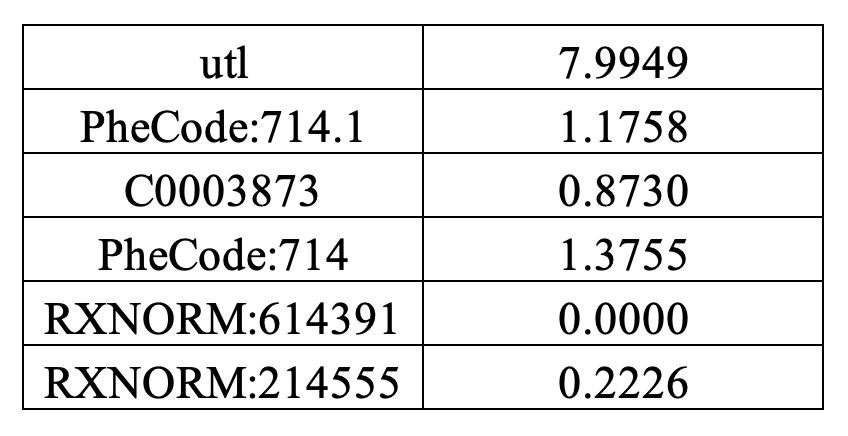
- Sheet 3: Conditional mean vector among patients with negative disease status.
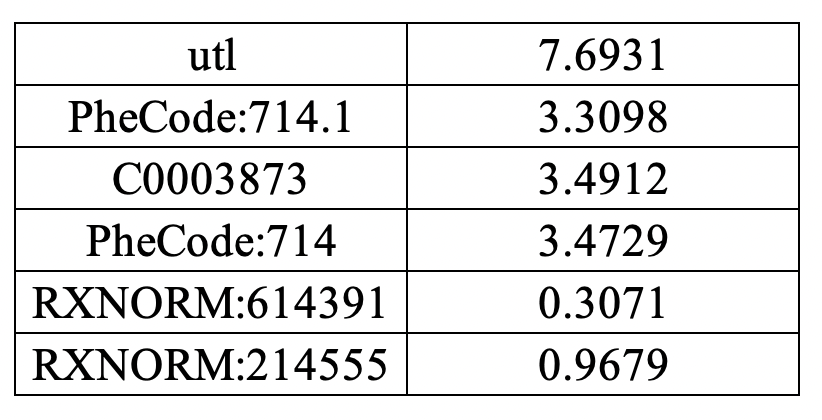
- Sheet 4: Conditional mean vector among patients with positive disease status.
- Sheet 5: A single number indicating the disease prevalence.
Sheet 1:
Sheet 2:
Sheet 3:
Sheet 4:
Covariance matrices (train + valid)
Dictionary
Feature names
Conditional suammry data
Model outcomes:
